Comparison with other models
compare.RmdThis vignette shows a brief comparison of the survival_ln_mixture
with other survival models, available through censored.
We begin by loading the packages and preparing the data.
library(lnmixsurv)
library(readr)
require(censored)
require(purrr)
require(dplyr)
require(ggplot2)
set.seed(4)
# Gerando dados
data <- simulate_data(n = 6000, k = 3, mixture_components = 2,
percentage_censored = 0.3)$data |>
filter(t < 500) |>
rename(x = cat, y = t)
new_data <- data |>
distinct(x)
formula <- Surv(y, delta) ~ xFor comparison, lets also estimate the Kaplan-Meier survival function.
library(ggsurvfit)
km <- survfit2(formula, data)
surv_km <- tidy_survfit(km, type = "surv") |>
select(.eval_time = time, .pred_survival = estimate, id = strata) |>
tidyr::nest(.pred = c(.eval_time, .pred_survival))The we build our parsnip specifications and store them
in a list.
ln_survival <- survival_reg(dist = "lognormal") |>
set_engine("survival")
ph_survival <- proportional_hazards() |>
set_engine("survival")
decision_tree <- decision_tree(cost_complexity = 0) |>
set_engine("rpart") |>
set_mode("censored regression")
ln_mixture <- survival_reg() |>
set_engine("survival_ln_mixture",
iter = 4000, warmup = 2000, starting_seed = 10,
em_iter = 450, mixture_components = 3)
ln_mixture_em <- survival_reg() |>
set_engine("survival_ln_mixture_em",
iter = 250, starting_seed = 15,
mixture_components = 3)
specs <- list(
ln_survival = ln_survival, ph_survival = ph_survival, ln_mixture = ln_mixture, decision_tree = decision_tree, ln_mixture_em = ln_mixture_em
)Finally, thanks to the great parsnip API, we can fit and predict all models at once.
set.seed(1)
models <- map(specs, ~ fit(.x, formula, data))
pred_sob <- map(models, ~ predict(.x, new_data,
type = "survival",
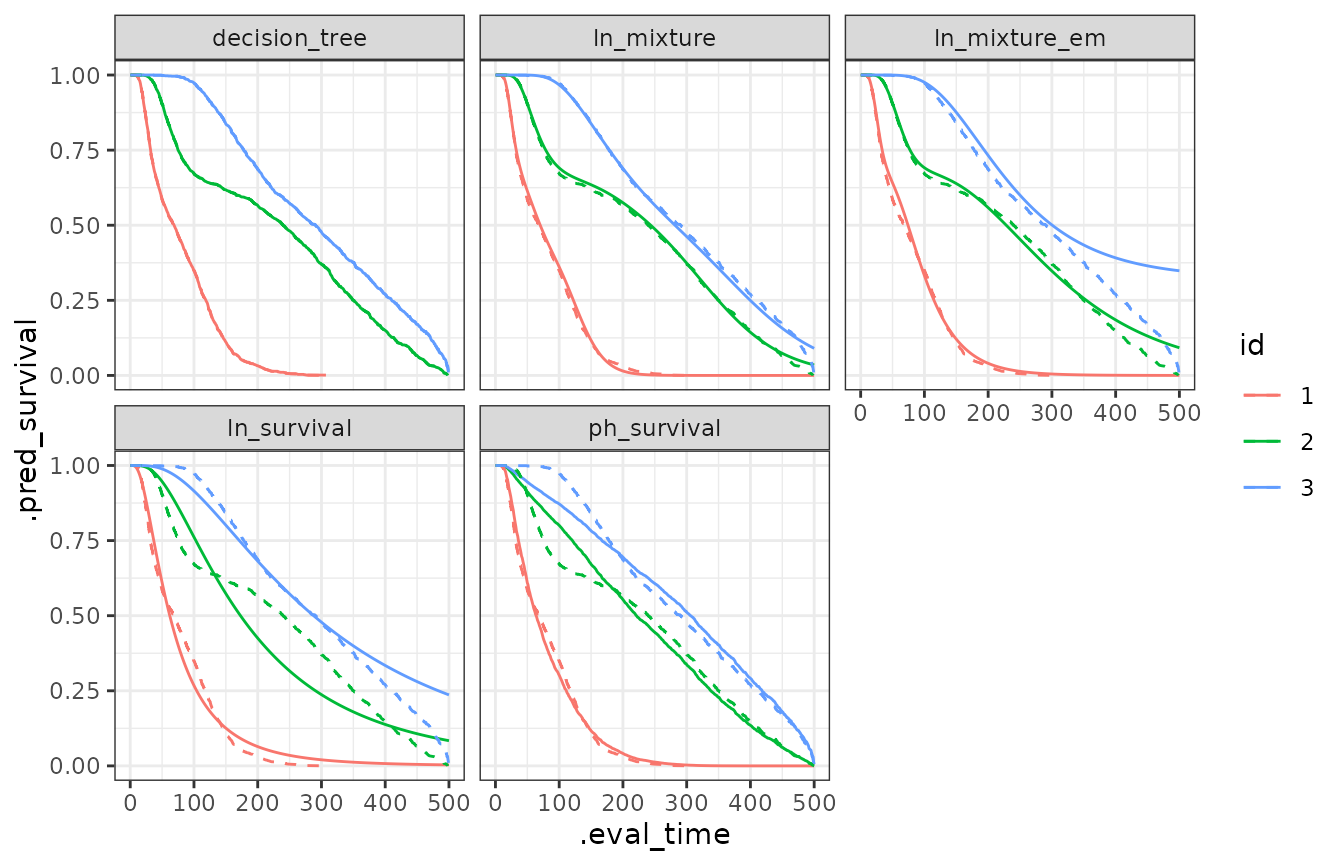
eval_time = seq(500)))The following plot compares each model with the Kaplan-Meier estimates of the survival function.
all_preds <- bind_rows(pred_sob, .id = "modelo") |>
group_by(modelo) |>
dplyr::mutate(id = new_data$x) |>
ungroup() |>
tidyr::unnest(cols = .pred)
km_fit <- surv_km |>
tidyr::unnest(cols = .pred) |>
filter(.eval_time < 500)
ggplot(aes(x = .eval_time, y = .pred_survival, col = id), data = all_preds) +
theme_bw() +
geom_line() +
facet_wrap(~modelo) +
geom_line(data = km_fit, linetype = "dashed")